Imaging and mapping the tumor-immune battlefield
Among the ever-evolving battle lines between the tumor and the immune system, what drives a durable and complete response to immunotherapy vs a partial response? How does the race between tumor growth and killing change with therapy? And how do we maximize this killing rate through positioning of the diverse units of the immune system? We aim to address these questions using engineered in vivo reporters, 3D tissue imaging, image analysis, spatial transcriptomics and mathematical modeling.
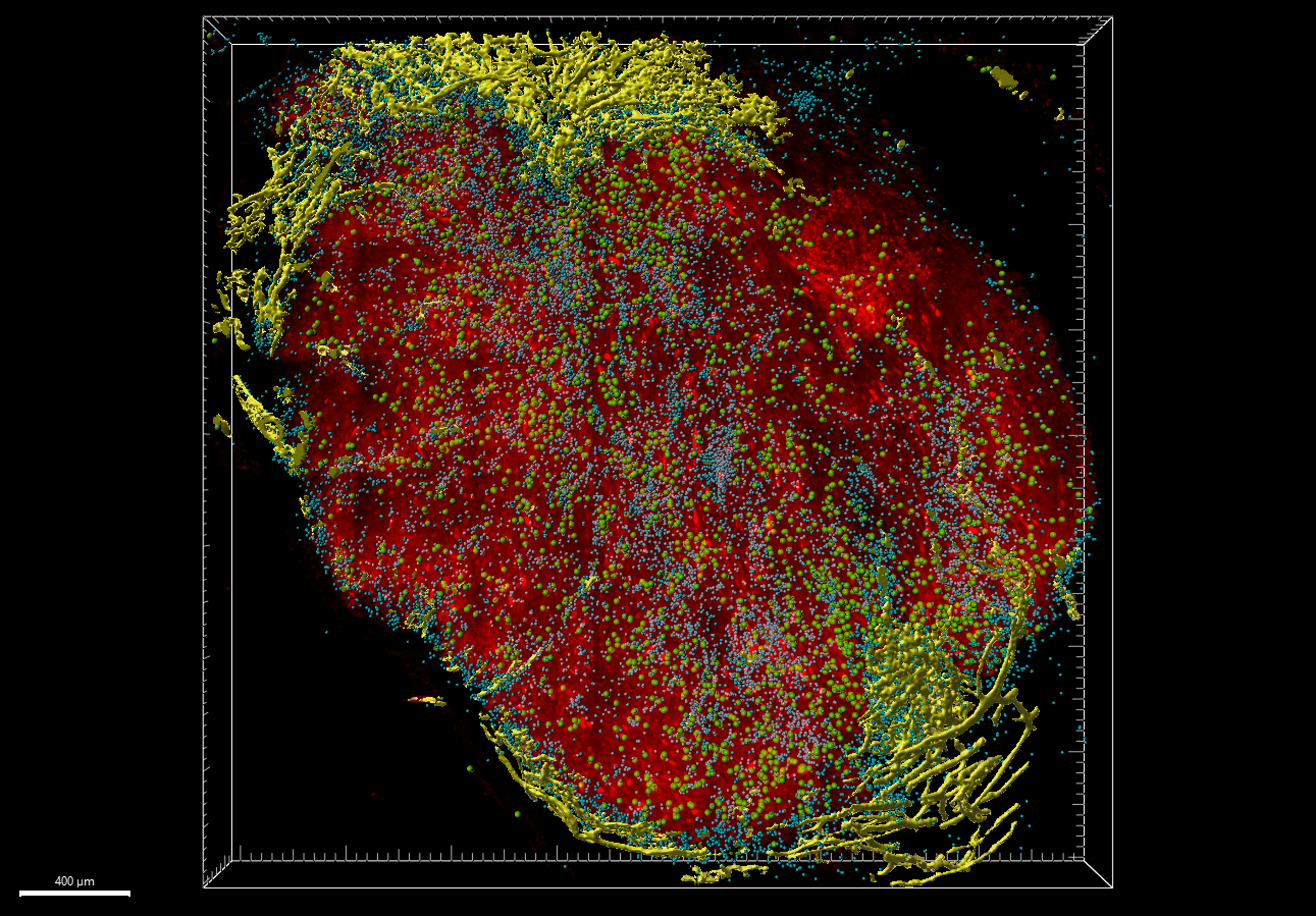
cleared 250 micron section of a mouse MC-38 tumor denoting tumor cells, CD8+ T cells, vasculature and dying tumor cells
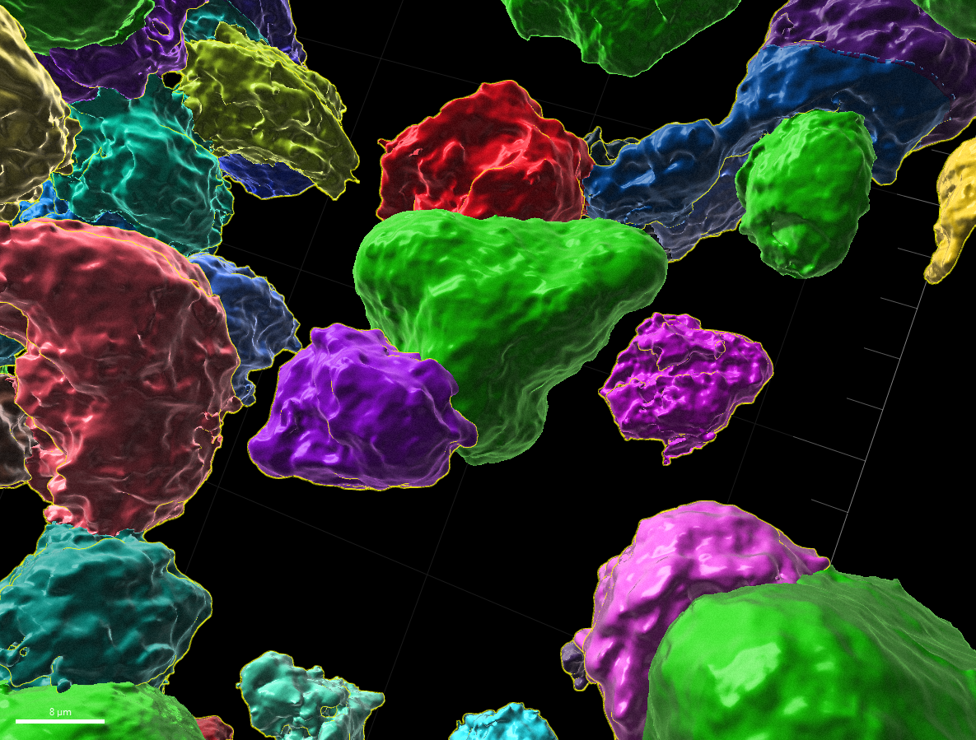
CD8+ T cells (assorted colors) attaching to dying tumor cells (green)
Dissecting and engineering the non-immune tumor stroma
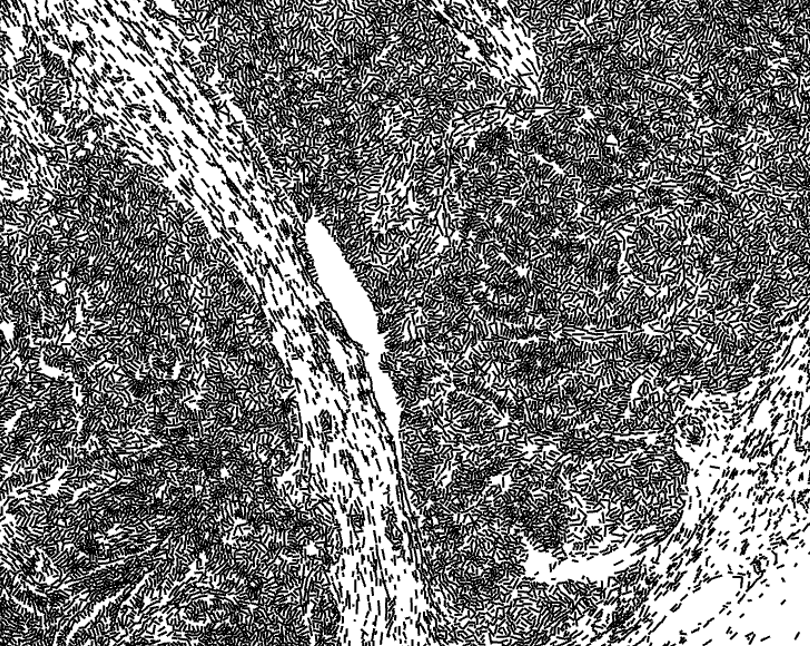
Blood vessels are the gateways to the tumor for cells and drugs. Distinct neighborhoods of cells develop along these routes. The 3D structure of the vascular network is intimately tied to it’s function, which mean we need to take our imaging to the 3rd dimension. How does the vascular network influence local cell states and vice versa? We use spatial transcriptomics, imaging and in vivo genetic perturbation tools to tease apart this network.
Fibroblasts can exert significant effects on the immune compartment of the tumor including ECM deposition, production of cytokines and direct antigen presentation. How are these diverse gene programs regulated by fibroblast signaling networks and how can we alter these networks to reprogram fibroblasts into anti-tumor states? We use single cell -omics and in vivo genetic perturbation tools to formulate and test hypotheses to address these questions.
Getting more bang for your buck when it comes to imaging:
With the explosion of high-plex imaging of proteins and transcriptomes, the amount of imaging data we are collecting as a field is growing rapidly. How much more biology can be learned if we look a little deeper into these images? We apply and develop image analysis algorithms to tie together cellular state, neighborhood, and morphology.
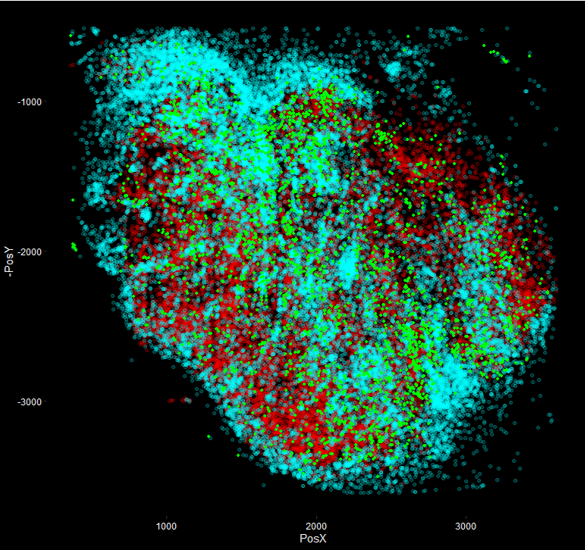
The field of spatial transcriptomics is rapidly growing, seeking to link the 3 dimensions of space with the many thousands of genes in transcriptomics space. We seek to utilize newly developed approaches such as ZipSeq to tackle questions around spatial tissue heterogeneity and how it arises and influences local immune cell activity.
We also seek to build on these approaches, adding new modalities, increasing their throughput with automation, increasing their resolution, as well as creating entirely new techniques integrating new photochemistry and genetic tools.